 On the surface, amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease share two commonalities: Both are progressively debilitating neurodegenerative conditions—meaning symptoms get worse—and, at least for now, neither has an effective treatment, let alone a cure.
On the surface, amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease share two commonalities: Both are progressively debilitating neurodegenerative conditions—meaning symptoms get worse—and, at least for now, neither has an effective treatment, let alone a cure.
On the molecular level, a recently identified connection between these devastating diseases could help advance the search for new therapies, according to Jeetain Mittal, the Sam and Ruth Madrid Endowed Chair in Chemical and Biomolecular Engineering.
Mittal and Brown University collaborator Nicolas Fawzi are focused on TAR DNA-binding protein 43, or TDP-43. Neuronal deposits of this essential human protein are found in people living with Alzheimer’s and related types of dementia, as well as those diagnosed with ALS (Lou Gehrig’s disease).
The research team has been awarded a $3.3 million grant from the National Institutes of Health to use their combined expertise in structural biology techniques—Fawzi’s is experimental; Mittal’s, computational—to visualize the atomistic details of TDP-43 assembly, explore the role of post-translational modification and disease-associated mutations on the assembly processes, and investigate interactions with several promising therapeutic targets to prevent aggregation.
The results of Mittal and Fawzi’s earlier work on TDP-43 have been published in Proceedings of the National Academy of Sciences and Structure. “Our previous research is only the tip of the iceberg,” says Mittal. “The next step is to take a look at the actual mutations associated with ALS and Alzheimer’s and get to the bottom of how they affect the assembly of TDP-43 to give us a better understanding of what causes their malfunction. Then, we can support researchers developing new therapeutic strategies.”
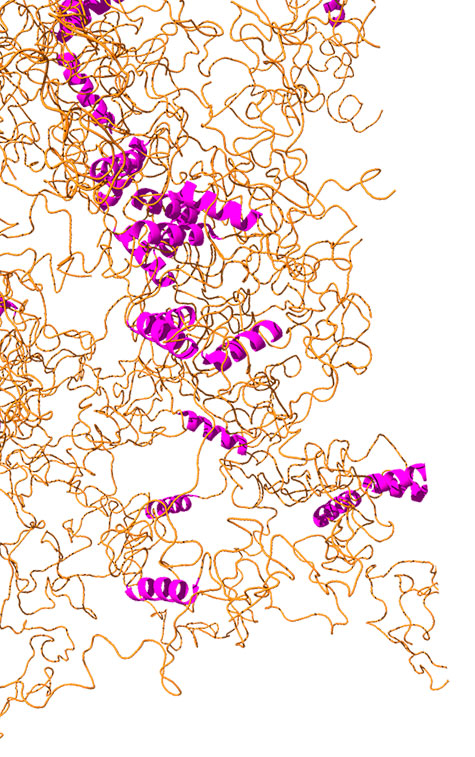
TDP-43 has two sides—one end of the molecule is made up of tightly packed folded regions, while the other side is classified as an intrinsically disordered region (IDR) because parts of it lack a well-defined structure. Mittal, a biophysicist, has a longstanding interest in IDRs and has developed simulation methods to help visualize how these proteins fold or misfold, potentially leading to disease-related aggregation in the body.
The unorganized nature of IDRs such as TDP-43 facilitates the formation of membrane-less organelles within cells through a process called liquid-liquid phase separation, a phenomenon that, according to Mittal, “has taken the biology world by storm.”
“Not all that long ago, people would think about a formation of organelles, or compartments within cells, the same way we’d think about a building having rooms separated with walls, and different functions happening in different rooms,” he explains. “The expectation was that the information within cells and the functions that take place inside of them were highly compartmentalized. But in the past 10 or 12 years, researchers have shown that cells do not require walls or membranes to make compartments.
“With liquid-liquid phase separation, you have a well-mixed solution of biomolecules, proteins, RNA, and so forth that suddenly coalesce in these very dense, liquid-like droplets. But if something goes wrong, and they don’t form properly, these granules become more solid-like and prone to aggregation.”
TDP-43 is a well-known component of these granules, he says, and because of the known genetic and other links to ALS, frontotemporal dementia, and Alzheimer’s disease, further study is of interest to the NIH.
The team is pursuing fundamental questions about the protein’s structure through two different but complementary approaches: Fawzi’s lab conducts highly sophisticated nuclear magnetic resonance experiments, confirming the protein’s loose, noodle-like structure through numerous readings averaged together. Mittal’s lab runs physics-based simulations that provide detailed spatial and time-related information and can predict how mutations to TDP-43 might affect phase behavior in the formation of membrane-less organelles.
Combining these methods allows the researchers “to get a mechanistic sense of how each step, from the atoms to the molecules to the single-protein level, connects,” Mittal says.
“Changing just a few atoms out of hundreds or thousands within the protein can affect its function, assembly, and interactions, so having a complete picture is important. Otherwise you’re just throwing everything at it and seeing what works. We’re trying to gain a better understanding so we can target our approach in a predictive manner.”
(ILLUSTRATION CREDIT: Biophysics and Soft Matter Group at Lehigh University)

