By using machine learning to analyze data from carbon nanotubes wrapped in DNA, researchers can detect both ovarian cancer biomarkers and actual cancer cells. (Illustration courtesy Daniel Heller/MSKCC)
Ovarian cancer kills 14,000 women in the United States every year. It’s the fifth leading cause of cancer death among women, and it’s so deadly, in part, because the disease is hard to catch in its early stages. Patients often don’t experience symptoms until the cancer has begun to spread, and there aren’t any reliable screening tests for early detection.
A team of researchers is working to change that. The group includes investigators from Memorial Sloan Kettering Cancer Center (MSK), Weill Cornell Medicine, the University of Maryland, the National Institutes of Standards and Technology (NIST), and Lehigh.
 Two recent papers describe their advancements toward a new detection method for ovarian cancer. The approach uses machine learning techniques to efficiently analyze spectral signatures of carbon nanotubes to detect biomarkers of the disease and to recognize the cancer itself.
Two recent papers describe their advancements toward a new detection method for ovarian cancer. The approach uses machine learning techniques to efficiently analyze spectral signatures of carbon nanotubes to detect biomarkers of the disease and to recognize the cancer itself.
The first paper appeared in Science Advances in November 2021.
“We demonstrated that a perception-based nanosensor platform could detect ovarian cancer biomarkers using machine learning,” says Yoona Yang, a postdoctoral research associate in Lehigh’s Department of Chemical and Biomolecular Engineering and co-first author along with Zvi Yaari, a postdoctoral research fellow at MSK. The authors also included Ming Zheng, a research chemist at NIST; Anand Jagota, a professor of bioengineering and chemical and biomolecular engineering and vice provost for research at Lehigh; and Daniel Heller, associate member and head of the Cancer Nanotechnology Laboratory at MSK.
“Perception-based sensing functions like the human brain,” says Yang. “The system consists of a sensing array that captures a certain feature of the analytes in a specific way, and then the ensemble response from the array is analyzed by the computational perceptive model. It can detect various analytes at once, which makes it much more efficient.”
For this study, the array consisted of single-wall carbon nanotubes wrapped in strands of DNA. The way in which the DNA was wrapped, and the variety of DNA sequences that were used, created a diversity of surfaces on the nanotubes. The diverse surfaces, in turn, attracted a range of proteins within a uterine lavage sample enriched with varying levels of ovarian cancer biomarkers.
“Carbon nanotubes have interesting electronic properties,” says Heller. “If you shoot light at them, they emit a different color of light, and that light’s color and intensity can change based on what’s sticking to the nanotube. We were able to harness the complexity of so many potential binding interactions by using a range of nanotubes with various wrappings. And that gave us a range of different sensors that could all detect slightly different things, and it turned out they responded differently to different proteins.”
The machine learning algorithm was trained using the data from the nanotube emission—the spectral signatures—to recognize the pattern of emission that signaled the presence and concentration of each biomarker.
“The mental breakthrough here is that these nanotubes are nonspecific sensors,” says Jagota. “They don’t know anything about biomarkers, meaning they aren’t programmed to bind to anything specific. All we knew is that they can be exposed to an aqueous medium, and whatever they’re exposed to within that medium will produce spectral shifts and changes in magnitude. And using a combination of these sensors, we were able to train the algorithm to mathematically transform these inputs to outputs with high accuracy.”
The second paper appeared in March 2022 in Nature Biomedical Engineering and comprised the work of many of the same researchers.
“In this paper, we weren’t looking at biomarkers any longer, we were looking at the disease itself,” says Heller. “We wanted to know, could this technology differentiate a blood sample from a patient with ovarian cancer from a patient without ovarian cancer?”
Those patients without ovarian cancer included both healthy people and people with other diseases.
In this study, the nanotubes were functionalized with quantum defects, which essentially increased the diversity of responses the nanotubes would provide.
“The nanotubes had a certain molecule bound to it that gave it an extra signal in terms of data,” says Jagota. “So richer data came from every nanotube-DNA combination. And the model was trained not on the biomarker, but on the disease state.”
The model developed a “disease fingerprint” from the spectral emissions of the nanotubes. The results were statistically significant in terms of the model’s specificity in detecting ovarian cancer and sensitivity in detecting both known and unknown biomarkers of the disease.
The team has shown their technique can detect ovarian cancer better than the current methods, but it can’t yet identify early stages of the disease. In part, says Heller, the issue is finding enough samples to train the algorithm because so few people are diagnosed at those time points.
“We’re working on determining how we can actually detect this disease at the earliest possible stages,” he says.
Next steps could also include branching out to develop the technique for a range of diseases, and determining if it can be optimized to work in clinical conditions, says Jagota.
Credits: Nature Biomedical Engineering Cover Image: Olga Kharchenko; Cover design: Alex Wing
A figure from the team's paper published in Science Advances illustrates "perception-based nanosensor platform for protein biomarkers."
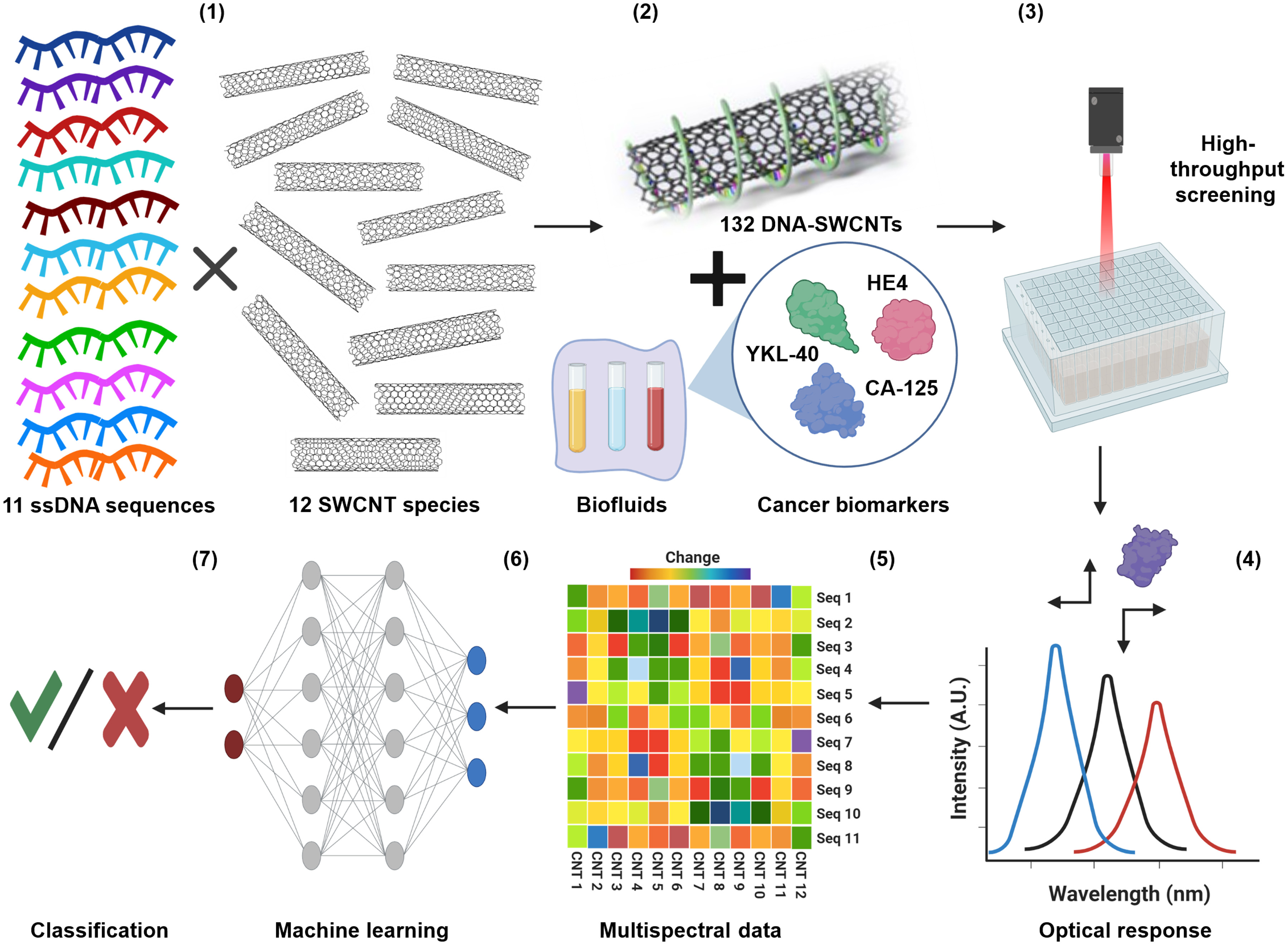
Caption: (1) Eleven single-stranded DNA oligonucleotides wrap SWCNT chiralities to form DNA-SWCNT sensor complexes. (2) The array of sensors is incubated in the sample of interest. (3) The optical response of the sensors is interrogated by high-throughput NIR spectroscopy. (4) The spectroscopic data are fitted to determine the wavelength and intensity of each sensor emission band. (5) The sensor responses are processed into a feature vector (FV) training set. A.U., arbitrary units. (6) ML algorithms are trained and validated for each target protein and their combinations. Seq, sequence; CNT, Carbon nanotubes. (7) Prediction results are evaluated.
Figure by Zvi Yaari
Yaari, Zvi & Yang, Yoona & Apfelbaum, Elana & Cupo, Christian & Settle, Alex & Cullen, Quinlan & Cai, Winson & Roche, Kara & Levine, Douglas & Fleisher, Martin & Ramanathan, Lakshmi & Zheng, Ming & Jagota, Anand & Heller, Daniel. (2021). A perception-based nanosensor platform to detect cancer biomarkers. Science Advances. 7. 10.1126/sciadv.abj0852.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).



